Histone modification (e.g., acetylation and methylation) is another major epigenetic mechanism in multicellular organisms that regulates gene expression and relates to disease etiology.
The basic structure unit of chromatin is nucleosomes. A single nucleosome contains a histone octamer core, which is formed by two molecules of each of the four core histones (H2A, H2B, H3 and H4) and ~146 bp DNA wrapped around this octamer core.
N-terminal histone tails are protruded from the nuclesome core and can be post-translationally modified by a number of processes, such as acetylatioin, methylaton, phosporylation. The covalent modification of histones constitutes a potential "histone code" that can be stably transmitted from parent cell to daughter cells.
The histone acetylation (Kac) has been extensively studied and shown to be closedly linked to gene transcriptional regulation through modified chromatin configuration and transcription factor binding. For example, histone acetylation at H3K9 (H3K9ac) and H4K12 (H4K12ac) have been demonstrated to be a predominant signal for gene activation by enhancing the accessibility of the transcription machinery, with the level of acetylation correlating with the rate of transcription. In contrast, histone deacetylases (HDACs) remove acetyl groups, which results in condensed chromatin and gene inactivation. H3K9ac and H4K12ac have been implicated as a key regulatory mechanism in the etiology of a variety of human complex diseases.
Histone acetylation status of several bone candidate genes has been studied in terms of their effects on gene expression and subsequent bone cell differentiation. For example, in vitro experiments indicated that the HDAC inhibitor, Trichostatin A (TSA), has a significant effect on the endogenous expression of RANKL (well known for its significance in bone metabolism) through enhanced acetylation of histones H3 and H4 on the proximal RANKL promoter sequence in bone stromal cells (Fan X, et al., J Cell Biochem., 2004. 93: 807-18). In addition, H3K9ac and H4ac were greatly enhanced at the promoters of osterix and osteocalcin (OC) genes in differentiated osteoblasts, and HDAC inhibitors can enhance osteoblast differentiation and osteogenic gene expression (Lee HW et al. 2006 Mol Endocrinol., 20:2432-43).
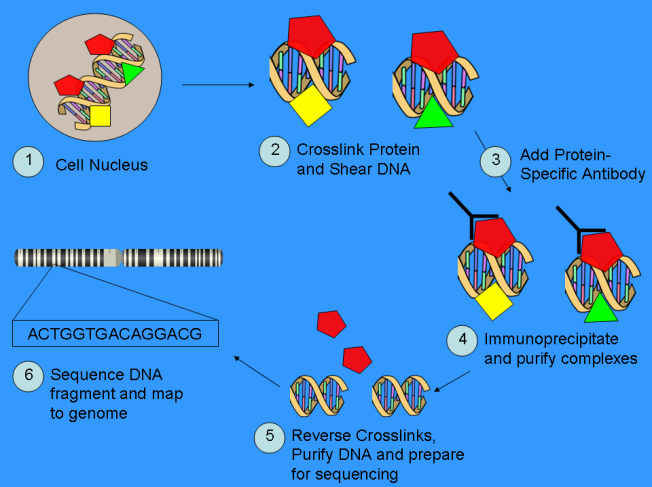
Despite these pioneering studies on cultured cell lines, the global histone modification profiles in primary human osteogenic cells related to risk of osteoporosis are largely unknown. We are initiating a pilot epigenome-wide H3K9ac and H4K12ac profiling analysis in human PBMs (potential precursors of bone-resorbing osteoclasts), by using the ChIP-Seq technique (chromatin immunoprecipitation followed by next-generation sequencing). Moreover, we will compare the PBM histone modification profiles between subjects with high vs. low BMD values in order to identify human genomic locations with differential intensity of histone modifications, termed differential histone modification site (DHMSs), that are associated with BMD variations. Characterizing DHMSs and thus the associated genes in relation to osteoporosis will provide novel insights into the etiologic and pathogenic mechanisms of osteoporosis.