Recent discoveries of microRNAs -- small, noncoding RNAs of average 22 nucleotides in length --have revealed a new class of epigenetic mechanisms of gene regulation. microRNAs can regulate gene expression at the post-transcriptional level by inducing RNA degradation through non-perfect base-pairing with their target mRNAs or by inhibiting protein translation.
Mature functional microRNAs are generated from long primary microRNA (pri-microRNA) transcripts. The pri-microRNAs, which usually contain a few hundred to a few thousand base pairs, are first processed into 70-nucleotide pre-miRNAs by Drosha and Pasha inside the nucleus. Pre-microRNAs are then transported into the cytoplasm by Exportin 5 and Ran-GTP and are further processed into small RNA duplexes by Dicer. The functional strand of the microRNA duplex is then assembled into the RNA-induced silencing complex (RISC). Finally, the microRNA guides the RISC to its mRNA target for translational repression or mRNA degradation.
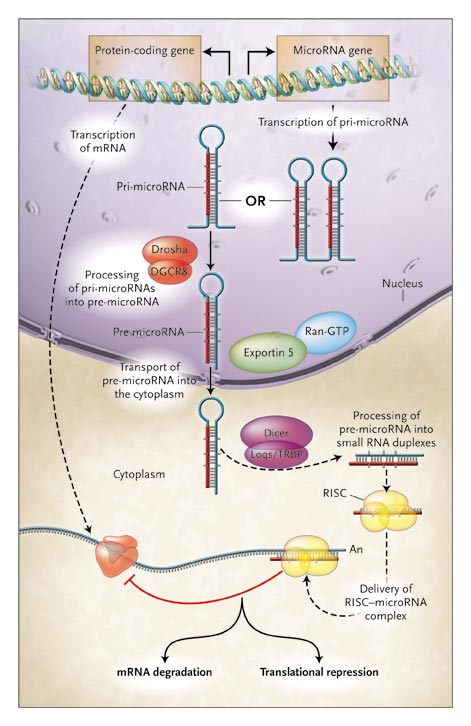
By Chen CZ, N Engl J Med. 2005; 353: 1768.
MicroRNAs have been implicated to play a profound role in an increasing number of human complex diseases such as cancer, Alzheimer's disease and cardiovascular diseases. The role of miRNAs in the pathophysiology of osteoporosis in humans is largely unknown at this time. Several studies have suggested that miRNAs play a significant role in osteogenic cell differentiation and bone metabolism.
For example, Sugatani et al. (J Cell Biochem. 2007; 101:996-9) demonstrated miR-223 isexpressed in mouse osteoclast precursor RAW264.7 cell line, and plays a key factor in osteoclast differentiation. A recent study by Li et al. (J Clin Invest. 2009; 119:3666-77) identified a new microRNA (miR-2861) in primary mouse osteoblasts that promotes osteoblast differentiation by repressing histone deacetylase 5 (HDAC5) expression at the post-transcriptional level. Importantly, miR-2861 was found to be conserved in humans, and a homozygous mutation in pre-miR-2861 that blocked expression of miR-2861 was shown to cause primary osteoporosis in 2 related adolescents. These results provided compelling evidence that microRNAs play important physiological roles in bone metabolism and pathogenesis of osteoporosis.
Recently we performed a pioneering microRNA profiling study of human PBMs and circulating B cells (another important osteoclastogenic cell) toidentify key microRNAs involved in BMD variation. Using ABI TaqManâLow Density Array that includes 365 human microRNA probes, we performed microRNA profiling of PBMs and B cells in 10 postmenopausal Caucasian females with high BMD vs. 10 with low BMD. In the PBMs, we identified a microRNA, miR-151, that was upregulated in the low vs. the high BMD subjects. By integrating the microRNA profiling data with mRNA expression profiling data of the same set of PBMs, we identified three genes that have expression levels correlated with that of miR-151, suggesting that they may be potential target genes of miR-151. The three genes are TNFSR11, LRCH1 and FZD5. In particular, the LRCH1 and FZD5 genes were also predicted as targets of miR-151 according to online microRNA bioinformatics analysis (http://www.targetscan.org). Our findings suggested that miR-151 may contribute to osteoporosis risk through regulating LRCH1 and FZD5. Similarly, we revealed that miR-181b is upregulated in B cells, in low vs. high BMD subjects. Further analysis on the genome-wide gene expression data in the same set of B cells suggested two potential target genes, FGFR1 and MECP2. Currently, we are performing a more comprehensive global microRNA profiling study for osteoporosis using high-density microRNA array and microRNA-Seq techniques, and are expected to identify additional novel microRNAs and their novel target genes for osteoporosis risk.

